Store multiple dataframes in a Python dictionary
If you have multiple pandas dataframes where you will do the same set of operations on each, rather than storing them in a list, store them in a dictionary.
A Python dictionary is data type built into Python that consists of a set of key: value pairs. Having this key can be very useful. It’s like a super simple contact book where the individual’s name is a key and the phone number is the value.
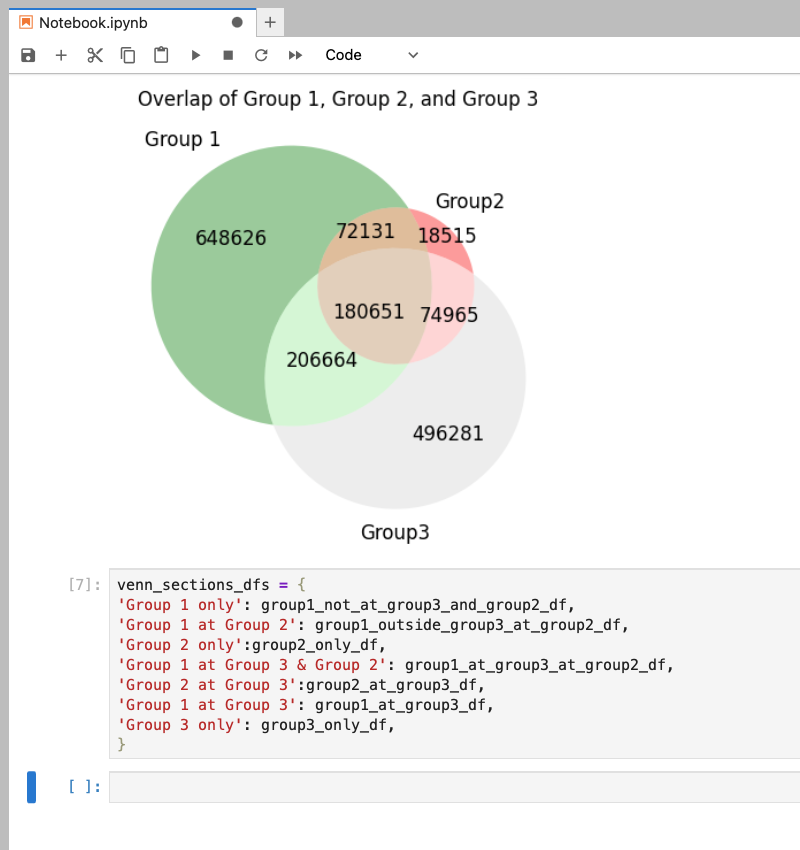
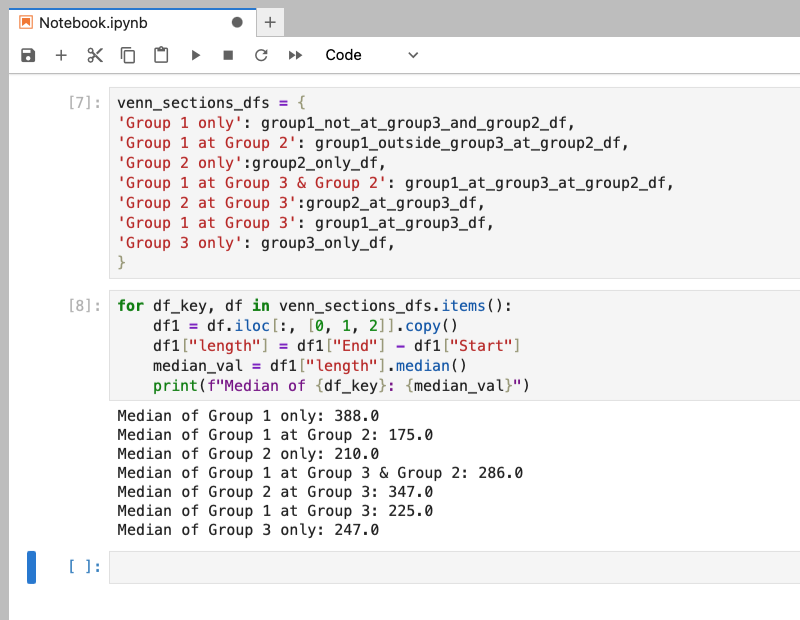
In regards to dataframes, our dictionary can consist of a string as the key and a dataframe as the value. This string can describe the dataframe it’s assigned to. In our examples here, the venn diagram represents common genomic regions between three samples. Our dictionary includes a key:value for each section of the venn diagram. So for the green section representing the regions unique to Group 1, the key is the string 'Group 1 only' and the value is the dataframe group1_not_at_group3_and_group2_df. This and the other pairs are stored in a dictionary called venn_sections_dfs. Having a description of our dataframe with our dataframe allows us to do a few tricks.
One thing we can do is print our keys in loops or functions. For example, our dataframes have a length column. If we wanted to know the average length of regions in each section of the venn diagram, we can loop through our dictionary, compute the median of the length column, and print the key with the calculated median value. It gives us a nice, human readable output as opposed to a list of medians.
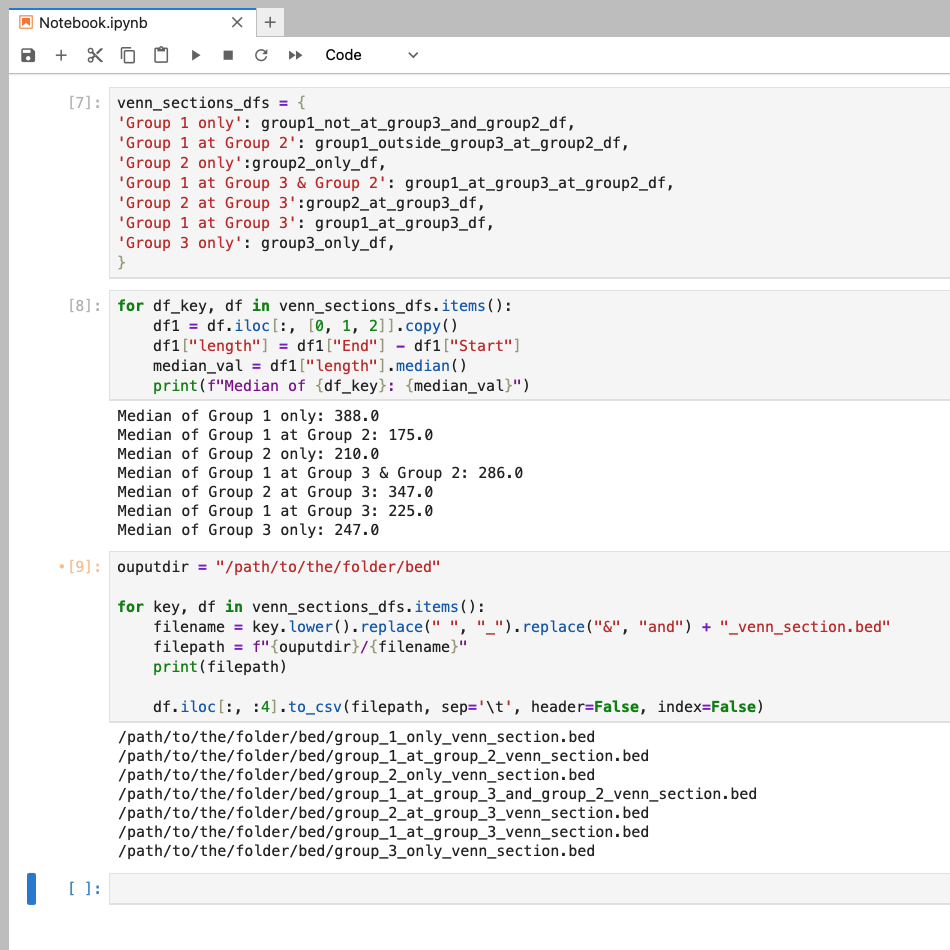
Another thing we can do, is use the key to create filenames. If we wanted to save our regions as a bed file, we can do this in a loop (assuming that all files will be saved to the same folder). However, instead of using the key as is, we are going to manipulate to make it better for working with on the HPC3 or the terminal in general. In our example, we need to replace spaces with _ and & with and. Optionally, we can add an extra string to our filename like _venn_section because we might be working with many bed files in our analysis. Add a print statement to check the new file name (and path) before you save your dataframes (make sure the line with to_csv is commented out, not like shown here). Finally, here we are only saving the first four columns by including iloc[:, :4].
In Summary
We used Python dictionaries to store and process multiple related dataframes where the keys describe the dataframes. More specifically, in our example, the keys were transformed to generate filenames and labels. You can easily add more items to the dictionary and write more complicated code that does not need to be duplicated for individual dataframes. Overall it makes your code more human readable, shorter, easier to maintain, and less error-prone.
Copy and Paste
You can copy and paste the following code blocks and update it for your code!
venn_sections_dfs = {
'Group 1 only': group1_not_at_group3_and_group2_df,
'Group 1 at Group 2': group1_outside_group3_at_group2_df,
'Group 2 only':group2_only_df,
'Group 1 at Group 3 & Group 2': group1_at_group3_at_group2_df,
'Group 2 at Group 3':group2_at_group3_df,
'Group 1 at Group 3': group1_at_group3_df,
'Group 3 only': group3_only_df,
}
for df_key, df in venn_sections_dfs.items():
df1 = df.iloc[:, [0, 1, 2]].copy()
df1["length"] = df1["End"] - df1["Start"]
median_val = df1["length"].median()
print(f"Median of {df_key}: {median_val}")
ouputdir = "/path/to/the/folder/bed"
for key, df in venn_sections_dfs.items():
filename = key.lower().replace(" ", "_").replace("&", "and") + "_venn_section.bed"
filepath = f"{ouputdir}/{filename}"
print(filepath)
#df.iloc[:, :4].to_csv(filepath, sep='\t', header=False, index=False)