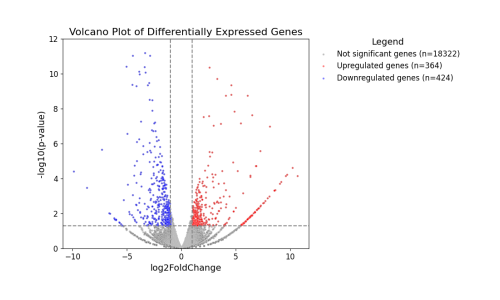
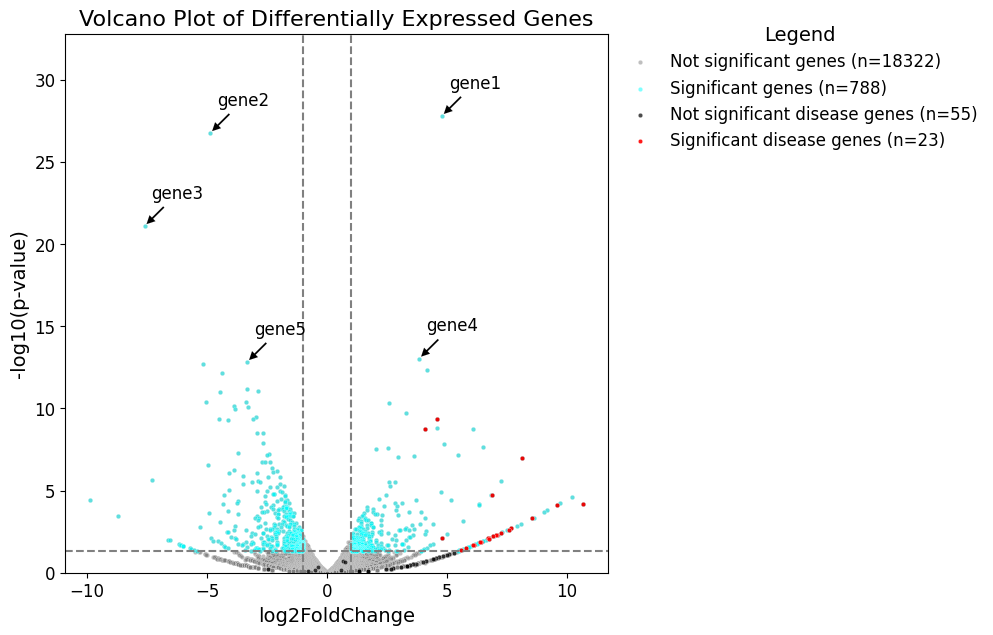
Plot a volcano plot with disease genes highlighted
This code looks for the columns 'gene_name', 'pvalue', 'log2FoldChange' in the dataframe.
For filtering data for a subset of genes, the defining regex patterns should look something like this:
subset_names_pattern = (
"gene1|gene2|gene3|gene4|gene5|gene6|gene7|"
"gene8|gene9|gene10|gene11|gene12|gene13|gene14|"
# more genes
"gene1000|gene1001|gene1002"
)
subset_drop_names_pattern = (
"geneA1|geneP2|geneP3|geneZ4|geneR5|geneD6|geneD7|"
"geneZ100|geneZ89|geneZ362"
)
The volcano plot:
import time
import pandas as pd
print("pandas: " + pd.__version__)
import numpy as np
print("numpy: " + np.__version__)
import seaborn as sns
print("seaborn: " + sns.__version__)
import matplotlib
import matplotlib.pyplot as plt
# embed plots in notebook
%matplotlib inline
print("matplotlib: " + matplotlib.__version__)
print(time.strftime("%Y-%m-%d"))
fig_title="Volcano Plot of Differentially Expressed Genes"
path = "/path/to/the/directory/for/plots"
fig_name=f'{path}/volcano.png'
print(fig_name)
# Define thresholds
pvalue=0.05
logfc = 1
n=5 # number of top genes to label
# Add -log10(p-value) column for plotting
degs_df["neg_log10_pval"] = -np.log10(degs_df["pvalue"])
# Filter for significant genes
pvalue_df = degs_df[(degs_df["pvalue"] <= pvalue) & (abs(degs_df["log2FoldChange"]) >= logfc)]
# Disease subset genes (by regex pattern)
degs_subset_df = degs_df[degs_df["gene_name"].str.contains(subset_names_pattern, regex=True, na=False)]
degs_subset_df = degs_subset_df[~degs_subset_df["gene_name"].str.contains(subset_drop_names_pattern, regex=True, na=False)]
pvalue_subset_df = degs_subset_df[(degs_subset_df["pvalue"] <= pvalue) & (abs(degs_subset_df["log2FoldChange"]) >= logfc)]
other_subset_df = degs_subset_df[~(degs_subset_df["pvalue"] <= pvalue)]
# Plotting
fig, ax = plt.subplots(figsize=(7, 8))
dot_size=10
# Plot each group of genes
group0_title=f'Not significant genes (n={len(degs_df)})'
sns.scatterplot(data=degs_df, x='log2FoldChange', y='neg_log10_pval', alpha=0.5, color='grey', label = group0_title, s=dot_size)
group1_title=f'Significant genes (n={len(pvalue_df)})'
sns.scatterplot(data=pvalue_df, x='log2FoldChange', y='neg_log10_pval', alpha=0.5, color='cyan', label = group1_title, s=dot_size)
group2_title=f'Not significant disease genes (n={len(other_subset_df)})'
sns.scatterplot(data=other_subset_df, x='log2FoldChange', y='neg_log10_pval', alpha=0.7, color='black', label = group2_title, s=dot_size)
group3_title=f'Significant disease genes (n={len(pvalue_subset_df)})'
sns.scatterplot(data=pvalue_subset_df, x='log2FoldChange', y='neg_log10_pval', alpha=0.9, color='red', label = group3_title, s=dot_size)
plt.xlabel(f'log2FoldChange', fontsize=14)
plt.ylabel(f'-log10(p-value)', fontsize=14)
ax.tick_params(axis='both', which='major', labelsize=12)
plt.title(fig_title, fontsize=16)
plt.legend(bbox_to_anchor=(1, 0.9), loc='center left',
title="Legend", title_fontsize=14, fontsize=12, frameon=False) # move legend
# loc options: 'best', 'upper right', 'upper left', 'lower left', 'lower right',
# 'right', 'center left', 'center right', 'lower center', 'upper center', 'center'
# draw horizontal and vertical dashed lines
yline = -np.log10(pvalue)
plt.axhline(yline, color='grey', linestyle='--')
plt.axvline(logfc, color='grey', linestyle='--')
plt.axvline(-logfc, color='grey', linestyle='--')
# Top N most significant genes
top_genes = degs_df.sort_values("pvalue").head(n)
# Label the top genes
for i, (idx, row) in enumerate(top_genes.iterrows()):
ax.annotate(
row["gene_name"],
xy=(row["log2FoldChange"], row["neg_log10_pval"]),
xytext=(5, 20),
textcoords="offset points",
fontsize=12,
color="black",
arrowprops=dict(arrowstyle="simple,head_length=0.5,head_width=0.5,tail_width=0.1",
color="black", lw=0.05)
)
# Set max for y-axis
plt.ylim(0, degs_df["neg_log10_pval"].max() + 5)
# Save the plot
plt.savefig(fig_name, bbox_inches = 'tight', format = 'png', dpi=300)
plt.show()